Welcome to pixStem’s documentation!¶
News¶
2021-2-1: pixStem has merged with pyXem!
To get a more robust software project for analysing pixelated STEM/4D-STEM/scanning electron diffraction data, pixStem has merged with pyXem. This means that all the functionality found in pixStem, is now available in pyXem. Some functions have been renamed and there has been some changes in how the functions work. So if you want to change to pyXem, some small modifications will be necessary for scripts or Jupyter Notebooks which used pixStem. Any bugs related to this can be reported at: pyXem issue tracker.
This webpage, and latest version of pixStem will remain available for the foreseeable future.
- pyXem: https://github.com/pyxem/pyxem/
- pyXem tutorials (Jupyter Notebooks): https://github.com/pyxem/pyxem-demos
2020-2-19: pixStem 0.4.0 released!
- Several functions for analysing low convergence STEM diffraction data, like nanobeam electron diffraction:
peak_position_refinement_com(),subtract_diffraction_background(),intensity_peaks()and a Laplacian of Gaussian (LoG) method tofind_peaks(). See Analysing NBED data or the Jupyter Notebook for an extended example. Thanks to Daen Jannis for the contribution! - Improvements to Template matching with a disk or ring and Peak finding, so they now work with any number of navigation dimensions, from single diffraction images, line scans and larger. This will be expanded to the other functions in the future, so they will allow for any data dimensionality.
- Add two new template matching functions: template_match_ring and Template matching with any binary image
- Added two new Jupyter Notebooks using data from journal articles with open data: magnetic characterization with 4-D STEM-DPC and structural characterization with 4-D STEM.
About pixStem¶
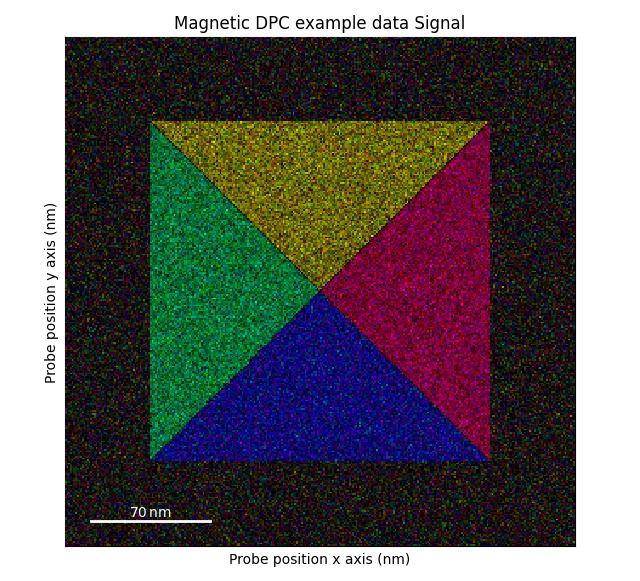
Library for processing data acquired on a fast pixelated electron detector, acquired using scanning transmission electron microscopy (STEM). For more information about this technique and how do analyse this type of data, see the paper Fast Pixelated Detectors in Scanning Transmission Electron Microscopy. Part I: Data Acquisition, Live Processing and Storage at arXiv.
Install instructions: Installing.
pixStem is available under the GNU GPL v3 license, and the source code is found at GitLab repository.
Contents:
Old news¶
2019-3-5: pixStem 0.3.3 released!
This release includes:
- Improvements to template_match_disk and Peak finding, so they now work with any number of navigation dimensions, from single diffraction images, line scans and larger. This will be expanded to the other functions in the future, so they will allow for any data dimensionality.
- Improved documentation for the fluctuation electron microscopy analysis. Thanks to Andrew Herzing for this contribution!
- Several minor bug-fixes.
2018-12-3: pixStem 0.3.2 released!
This release includes:
- Data analysis functionality for analysing fluctuation electron microscopy data, thanks to Andrew Herzing for the contribution!
- A new method in the
PixelatedSTEMclass for template matching a disk with STEM diffraction data: Template matching with a disk or ring - Another method for locating diffraction spots in STEM diffraction data, using peak finding functionality from
skimage: Peak finding
All these rely on the dask library, so they can be performed on very large datasets.
2018-6-28: pixStem 0.3.1 released!
This is a minor release which includes functionality for correcting both dead and hot pixels through find_dead_pixels(), find_hot_pixels() and correct_bad_pixels().
Featurewise it also includes a simple function for loading binary signals: pixstem.io_tools.load_binary_merlin_signal().
Unit testing of docstrings has been improved, with a combined setup and teardown function, and introduction of pixstem and numpy into the docstring’s namespace.
2018-6-13: pixStem 0.3.0 released!
fpd_data_processing has been renamed pixStem!
This release also includes greatly improved center of mass, virtual annular dark field and virtual bright field functions, which now uses only dask array operations.
This means they can return lazy signal, which makes it much easier to work with large datasets on computers with limited memory.
It also includes several backend improvements to DPCSignal.correct_ramp and utility functions for getting diffraction values.
A Jupyter Notebook giving a basic introduction to pixStem’s features is available at pixStem demos.
2018-2-18: fpd_data_processing 0.2.1 released!
This release includes a major improvement for lazy signal processing with center of mass and radial integration. It also includes a new method for shifting diffraction patterns in the PixelatedSTEM class.
Acknowledgement¶
Initial work from Magnus Nord funded by EPSRC via the project “Fast Pixel Detectors: a paradigm shift in STEM imaging” (Grant reference EP/M009963/1).